Characterization of cancer-associated fibroblasts (CAFs) and tumor-infiltrating lymphocytes (TILs) in non-small cell lung cancer
Chrysa Papadopoulou
Last updated: 2025-10-01
Checks: 7 0
Knit directory: CCL19_FRCs_lung_cancer/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240808) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 25e9eba. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/Final_submission/
Ignored: data/Human/
Ignored: data/Mouse/
Ignored: data/Public/
Ignored: output/GSEA_AdvFB_SULF1/
Ignored: output/GSEA_AdvFB_TLS/
Ignored: output/GSEA_CCR7_T/
Ignored: output/GSEA_CD8_T/
Ignored: output/GSEA_CYCL_T/
Ignored: output/GSEA_EXH_T/
Ignored: output/GSEA_SMC_PRC/
Untracked files:
Untracked: README.html
Untracked: analysis/.h5seurat
Untracked: analysis/Compare_tumors.Rmd
Untracked: analysis/NSCLC_PDAC_CAFs.Rmd
Untracked: analysis/QC_NSCLC.Rmd
Untracked: analysis/Seurat_to_SCE.Rmd
Untracked: analysis/comparison_CCL19_FRC_NSCLC_tonsil.Rmd
Untracked: analysis/compression.Rmd
Untracked: analysis/gene_metaprograms.Rmd
Untracked: analysis/index_hidden.Rmd
Unstaged changes:
Modified: analysis/extra_functions.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/FRC_TIL_landscape.Rmd) and
HTML (docs/FRC_TIL_landscape.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 25e9eba | Pchryssa | 2025-10-01 | FRC TIL landscape |
| html | 9458db6 | Pchryssa | 2025-10-01 | Build site. |
| Rmd | cb1c77d | Pchryssa | 2025-10-01 | FRC TIL landscape |
Load packages
suppressPackageStartupMessages({
library(here)
library(purrr)
library(Seurat)
library(dittoSeq)
library(CellChat)
library(harmony)
library(ggsci)
library(bigmds)
})Set directory
basedir <- here()Characterization of cancer-associated fibroblasts (CAFs) in NSCLC
Read Stroma cell data
NSCLC_data <- readRDS(paste0(basedir,"/data/Human/NSCLC_stroma_total.rds"))Subset on CAFs only
CAFs_NSCLC <- subset(NSCLC_data, cell_type %in% c("CAF2", "CAF1"))
#Do preprocessing again without re normalization
resolution <- c(0.1, 0.25, 0.4, 0.6, 0.8, 1., 1.2, 1.4, 1.6, 2., 2.2, 2.4)
CAFs_NSCLC <- FindVariableFeatures(CAFs_NSCLC, selection.method = "vst", nfeatures = 2000)
CAFs_NSCLC <- ScaleData(CAFs_NSCLC)
CAFs_NSCLC <- RunPCA(object = CAFs_NSCLC, npcs = 30, verbose = FALSE,seed.use = 8734)
CAFs_NSCLC <- RunTSNE(object = CAFs_NSCLC, reduction = "pca", dims = 1:20, seed.use = 8734)
CAFs_NSCLC <- RunUMAP(object = CAFs_NSCLC, reduction = "pca", dims = 1:20, seed.use = 8734)
CAFs_NSCLC <- FindNeighbors(object = CAFs_NSCLC, reduction = "pca", dims = 1:20, seed.use = 8734)
for(k in 1:length(resolution)){
CAFs_NSCLC <- FindClusters(object = CAFs_NSCLC, resolution = resolution[k], random.seed = 8734)
}Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9577
Number of communities: 6
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9283
Number of communities: 11
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9120
Number of communities: 13
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8958
Number of communities: 20
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8832
Number of communities: 21
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8708
Number of communities: 24
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8601
Number of communities: 26
Elapsed time: 6 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8505
Number of communities: 27
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8410
Number of communities: 29
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8245
Number of communities: 36
Elapsed time: 6 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8183
Number of communities: 36
Elapsed time: 6 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1278142
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8112
Number of communities: 42
Elapsed time: 6 secondsData integration with Harmony
experiment.harmonized <- RunHarmony(CAFs_NSCLC,
group.by.vars = c("patient"),
reduction = "pca", reduction.save = "harmony", plot_convergence = TRUE)
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
harmony_embeddings <- Embeddings(experiment.harmonized, 'harmony')
experiment.harmonized <- RunUMAP(experiment.harmonized, dims = 1:30,seed.use = 1753,reduction = "harmony")
experiment.harmonized <- FindNeighbors(experiment.harmonized, dims = 1:30,seed.use = 1753, reduction = "harmony")
resolution <- c(0.1, 0.15, 0.125, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45 , 0.5, 0.6, 0.8, 1., 1.2, 1.4, 1.6, 1.8, 2., 2.2, 2.5)
for(k in 1:length(resolution)){
experiment.harmonized <- FindClusters(object = experiment.harmonized, resolution = resolution[k], algorithm = 1,random.seed = 1753)
}Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9550
Number of communities: 6
Elapsed time: 10 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9404
Number of communities: 8
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9479
Number of communities: 7
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9285
Number of communities: 8
Elapsed time: 10 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9177
Number of communities: 8
Elapsed time: 10 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9081
Number of communities: 10
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9001
Number of communities: 11
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8912
Number of communities: 11
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8844
Number of communities: 12
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8774
Number of communities: 12
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8661
Number of communities: 13
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8475
Number of communities: 16
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8340
Number of communities: 21
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8223
Number of communities: 24
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8127
Number of communities: 27
Elapsed time: 8 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8033
Number of communities: 26
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.7946
Number of communities: 31
Elapsed time: 6 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.7866
Number of communities: 33
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.7782
Number of communities: 34
Elapsed time: 7 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 40477
Number of edges: 1311218
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.7667
Number of communities: 36
Elapsed time: 7 secondsCancer associated fibroblasts - CAFs subsets (Figure 3A)
cols<- pal_igv()(51)
names(cols) <- c(0:50)
palet <- cols[4:6]
names(palet) <- c("CAF2","CAF1")
DimPlot(experiment.harmonized, reduction = "umap", group.by = "cell_type", cols= palet)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2") + ggtitle("Cancer associated fibroblasts")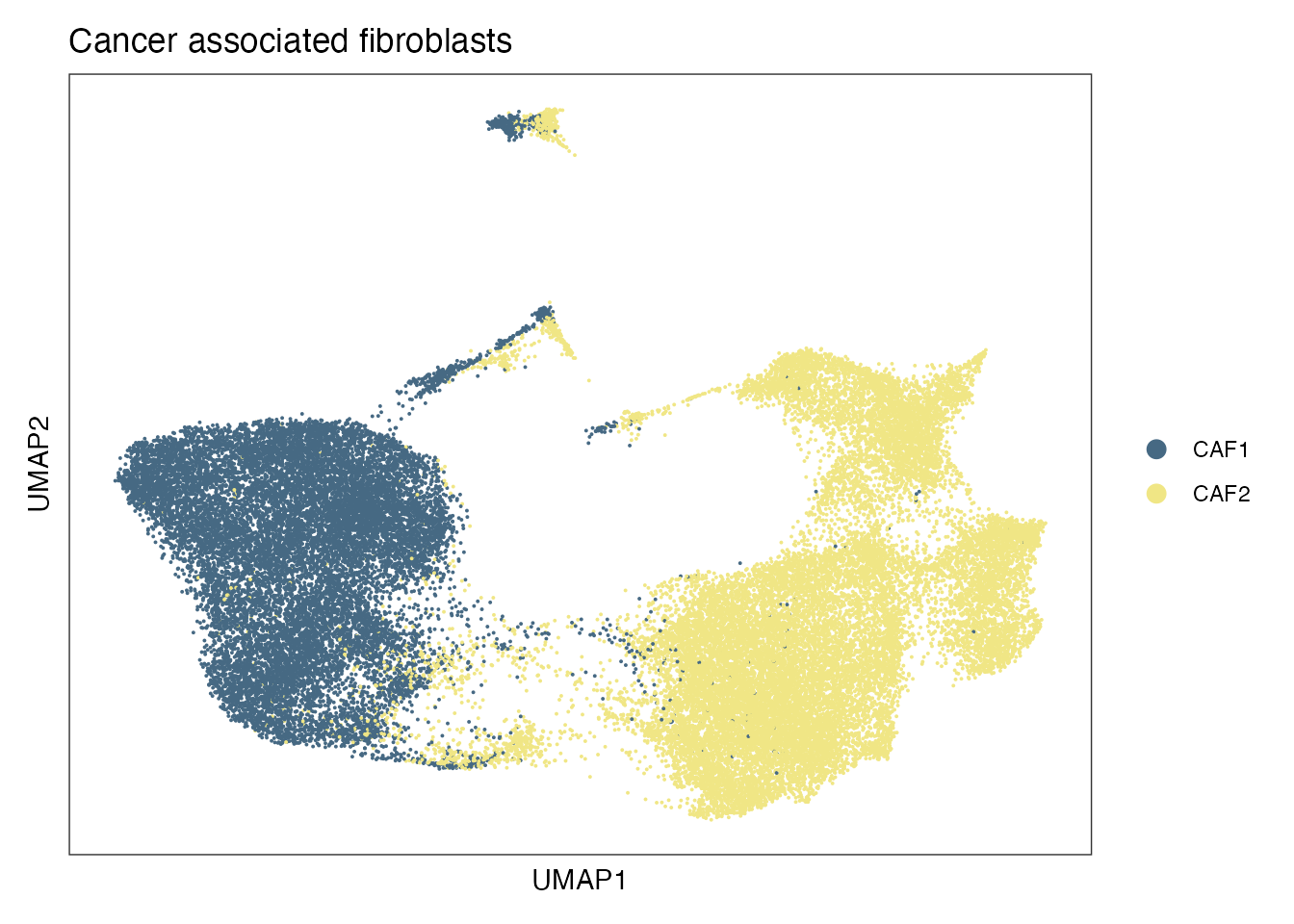
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
CCL19 expression in CAFs of NSCLC (Figure 4A)
FeaturePlot(experiment.harmonized, reduction = "umap",
features = get_full_gene_name('CCL19',experiment.harmonized),raster=FALSE,
cols=c("lightgrey", "red"))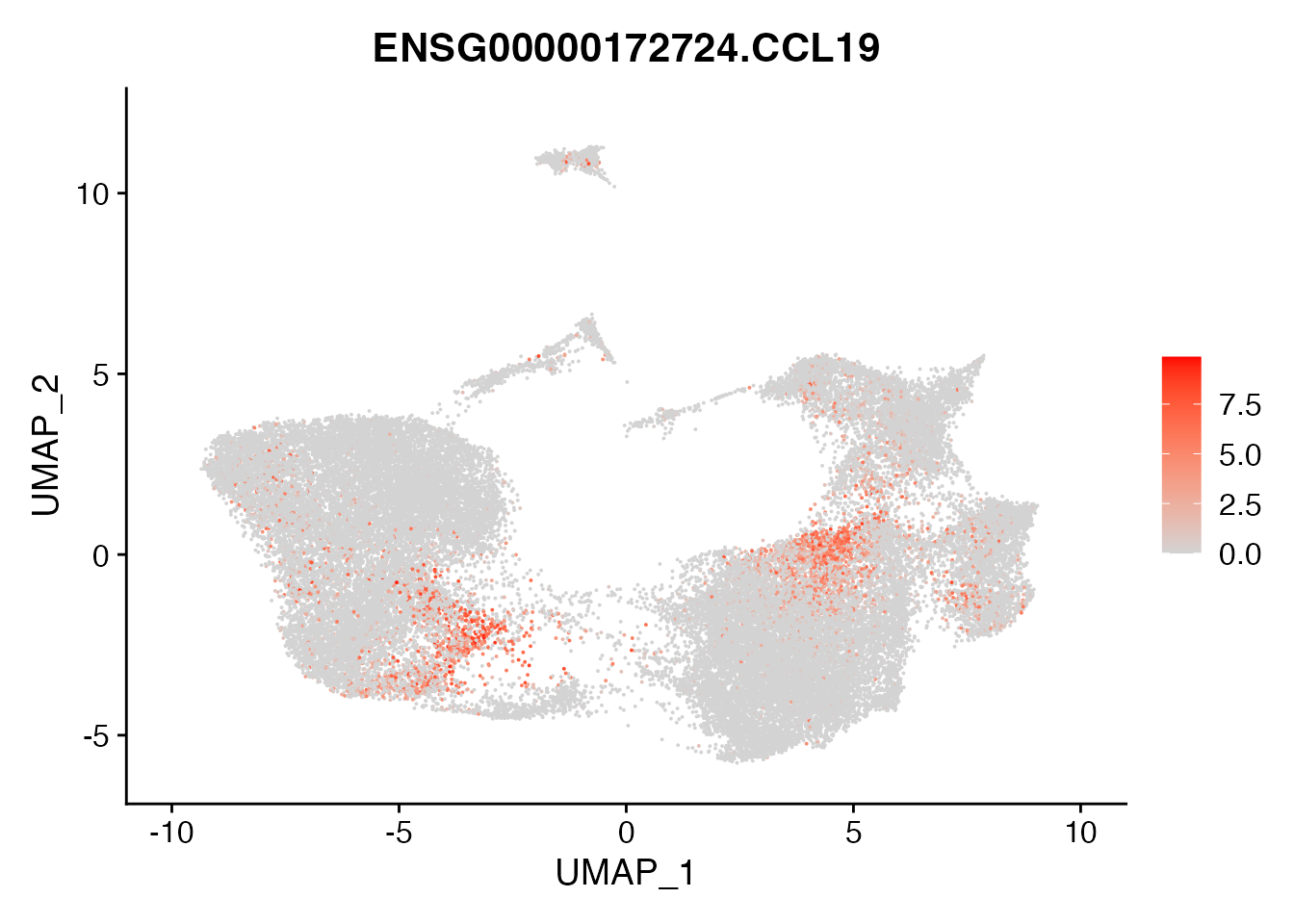
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
CAF gene signatures (Figure 3B)
data_conv <- experiment.harmonized
data_conv <-Remove_ensebl_id(data_conv)
gene_list <- c("CCL19","CCL21","PDPN","FAP","POSTN","CLU","LEPR","CD34","SULF1","DPT","ICAM1",
"VCAM1","CXCL12","ALDH1A1","MMP2","ACTA2","MYH11","MCAM","NOTCH3","RGS5",
"DES","AIFM2","CSPG4", "KCNJ8", "ITGA7")
dittoDotPlot(data_conv, vars = gene_list, group.by = "cell_type", size = 8,legend.size.title = "Expression (%)",scale = FALSE) + ylab(" ") 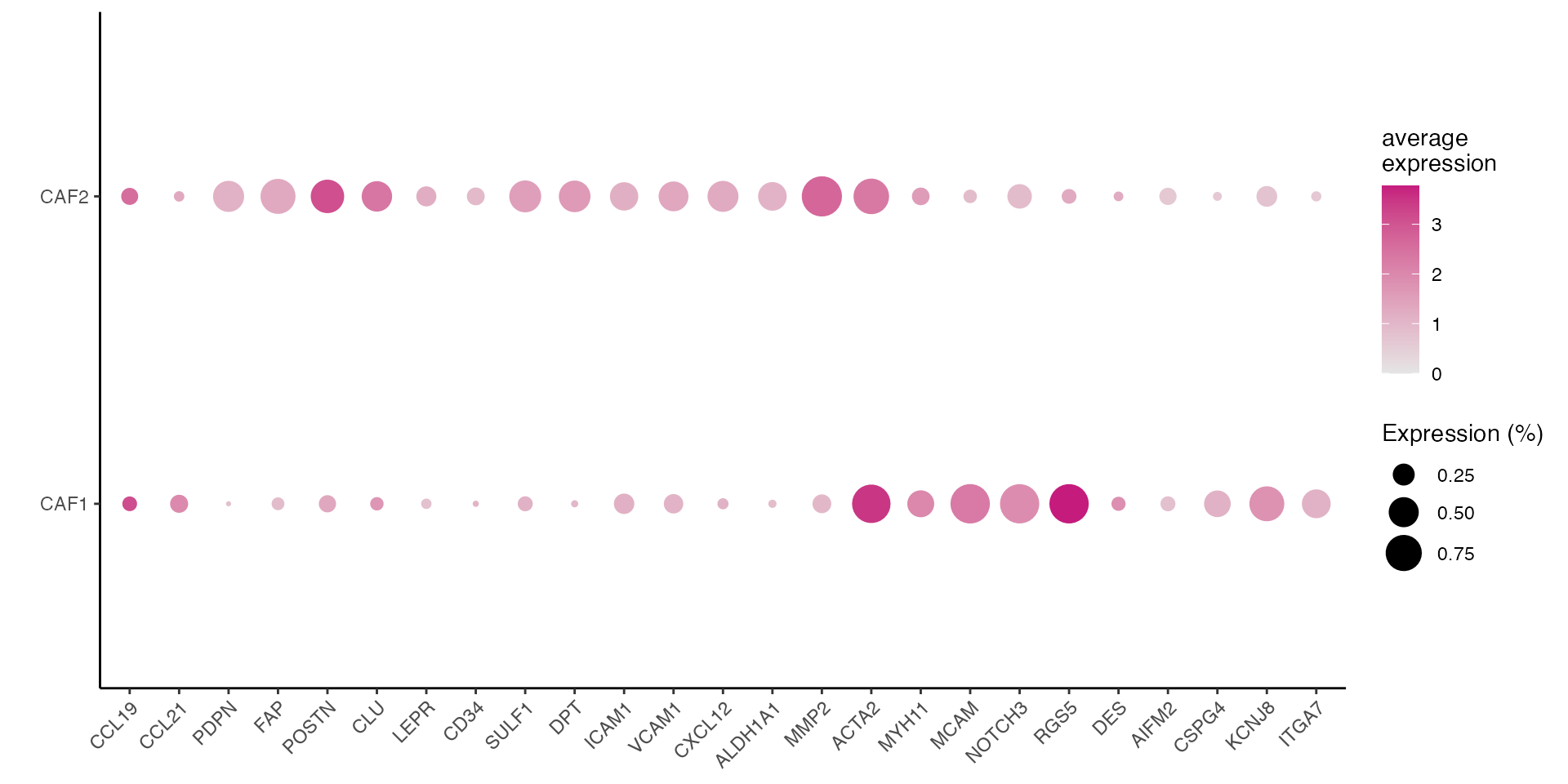
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
Characterization of tumor-infiltrating lymphocytes (TILs) in NSCLC
Read NSCLC TIL data
NSCLS_TIL_data <- readRDS(paste0(basedir,"/data/Human/NSCLC_TILs.rds"))Tumor-infiltrating lymphocytes (TILS) subsets (Figure 3C)
#Define color palet
palet <- c("#5050FFFF", "#CE3D32FF", "#4DAF4A","#FB9A99","#377EB8","#A65628","#222F75")
names(palet) <- c( "CAF2/TRC","CAF1/PRC", paste0("CD4", "\u207A ", "T cells"), paste0("CD8", "\u207A ", "T cells"), "B cells", "Regulatory T cells",paste0("Cycling CD8", "\u207A ", "T cells"))
DimPlot(NSCLS_TIL_data, reduction = "umap", group.by = "cell_type", cols=palet)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2") + ggtitle(paste0("Tumor infiltrating lymphocytes"))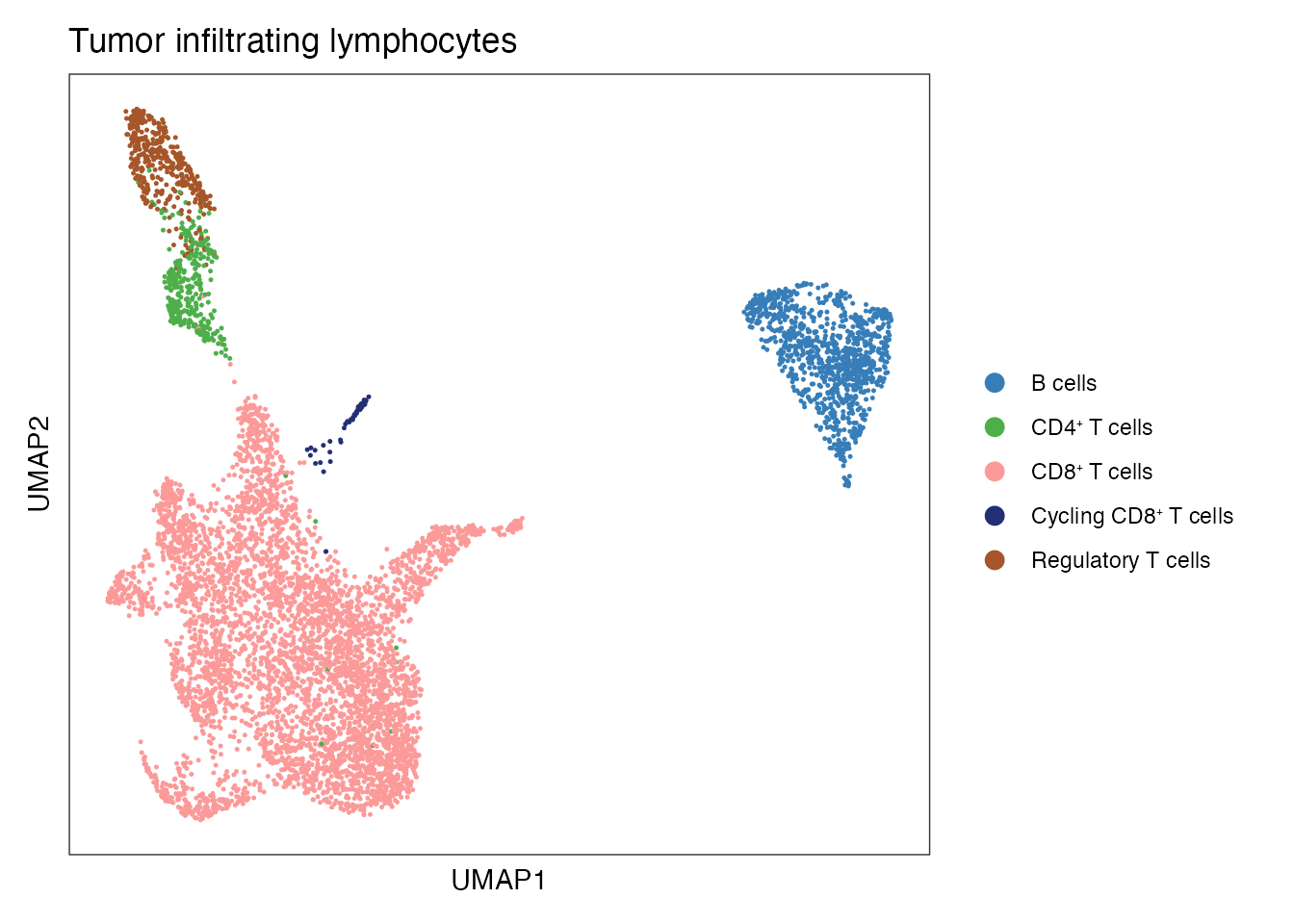
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
TIL gene signatures (Figure 3D)
data_conv <-Remove_ensebl_id(NSCLS_TIL_data)
genes <- c("CD79A","IGHM","IGHD","JCHAIN","CD4","CCR7","SELL","FOXP3","IKZF4","IL2RA","CTLA4","CD8A","CX3CR1","FCGR3A","KIR3DL2","KLRF1", "NKG7","GZMB","GNLY","TIGIT","TOP2A","MKI67","PCLAF")
data_conv$cell_type <- factor(data_conv$cell_type, levels = c("B cells",paste0("CD4", "\u207A ", "T cells"),"Regulatory T cells",paste0("CD8", "\u207A ", "T cells"), paste0("Cycling CD8", "\u207A ", "T cells")))
gg <-dittoDotPlot(data_conv, vars = genes, group.by = "cell_type", size = 4)
gg + coord_fixed(ratio=0.8) + ylab("")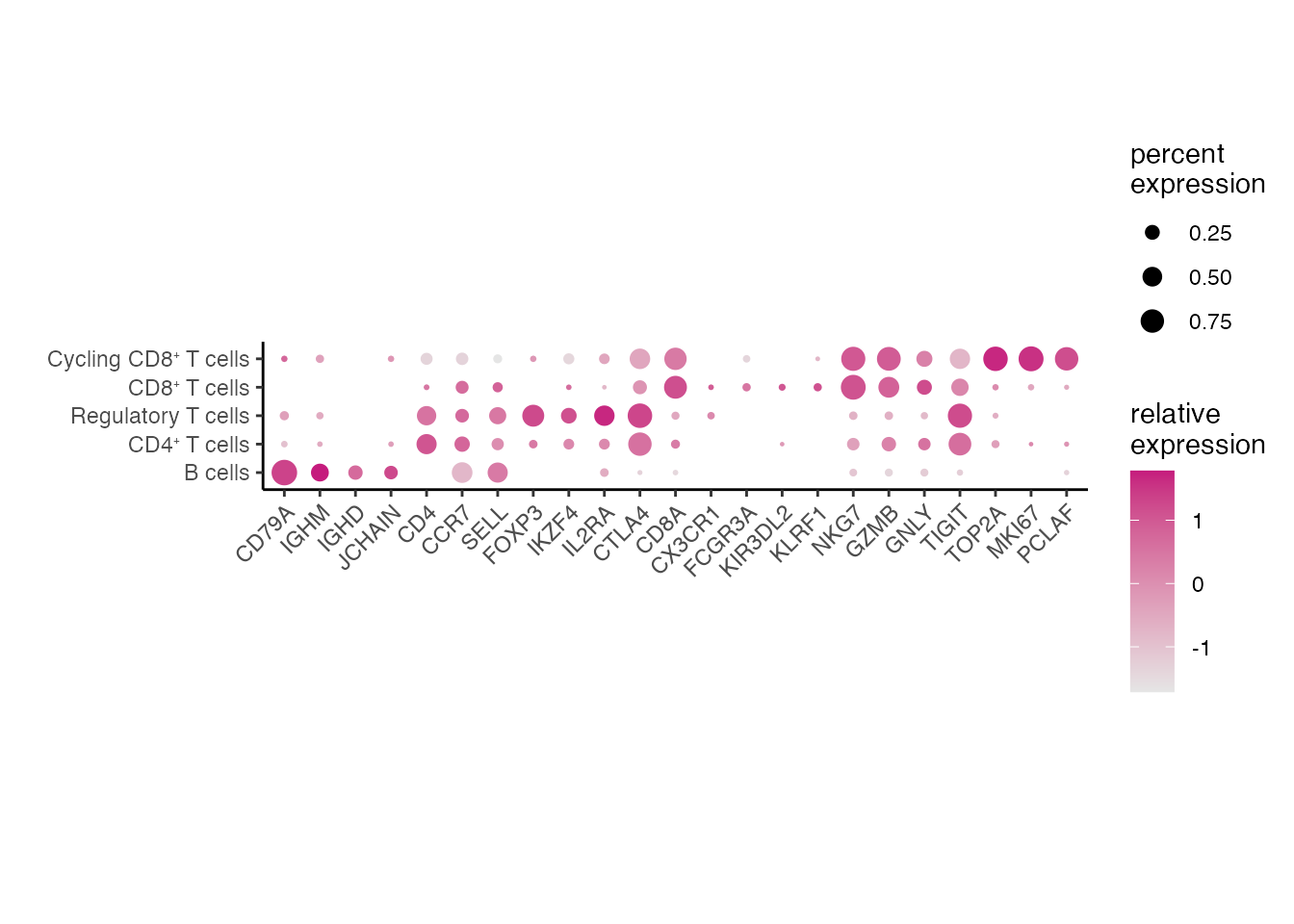
| Version | Author | Date |
|---|---|---|
| 9458db6 | Pchryssa | 2025-10-01 |
Session info
sessionInfo()R version 4.3.1 (2023-06-16)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.6.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] bigmds_3.0.0 ggsci_3.0.0 harmony_1.2.0
[4] Rcpp_1.0.11 CellChat_1.6.1 Biobase_2.60.0
[7] BiocGenerics_0.46.0 igraph_1.5.0.1 dplyr_1.1.2
[10] dittoSeq_1.12.1 ggplot2_3.4.2 SeuratObject_5.1.0
[13] Seurat_4.3.0.1 purrr_1.0.1 here_1.0.1
[16] magrittr_2.0.3 circlize_0.4.15 tidyr_1.3.0
[19] tibble_3.2.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.21 splines_4.3.1
[3] later_1.3.1 bitops_1.0-7
[5] polyclip_1.10-4 ggnetwork_0.5.12
[7] lifecycle_1.0.3 rstatix_0.7.2
[9] doParallel_1.0.17 rprojroot_2.0.3
[11] globals_0.16.2 processx_3.8.2
[13] lattice_0.21-8 MASS_7.3-60
[15] backports_1.4.1 plotly_4.10.2
[17] sass_0.4.7 rmarkdown_2.23
[19] jquerylib_0.1.4 yaml_2.3.7
[21] httpuv_1.6.11 NMF_0.26
[23] sctransform_0.4.1 spam_2.10-0
[25] sp_2.0-0 spatstat.sparse_3.0-2
[27] reticulate_1.36.1 cowplot_1.1.1
[29] pbapply_1.7-2 RColorBrewer_1.1-3
[31] abind_1.4-5 zlibbioc_1.46.0
[33] Rtsne_0.16 GenomicRanges_1.52.0
[35] RCurl_1.98-1.12 pracma_2.4.4
[37] git2r_0.33.0 GenomeInfoDbData_1.2.10
[39] IRanges_2.34.1 S4Vectors_0.38.1
[41] svd_0.5.5 ggrepel_0.9.3
[43] irlba_2.3.5.1 listenv_0.9.0
[45] spatstat.utils_3.1-0 pheatmap_1.0.12
[47] RSpectra_0.16-1 goftest_1.2-3
[49] spatstat.random_3.1-5 fitdistrplus_1.1-11
[51] parallelly_1.36.0 svglite_2.1.1
[53] leiden_0.4.3 codetools_0.2-19
[55] DelayedArray_0.28.0 tidyselect_1.2.0
[57] shape_1.4.6 farver_2.1.1
[59] matrixStats_1.0.0 stats4_4.3.1
[61] spatstat.explore_3.2-1 jsonlite_1.8.7
[63] GetoptLong_1.0.5 BiocNeighbors_1.18.0
[65] ellipsis_0.3.2 progressr_0.13.0
[67] ggalluvial_0.12.5 ggridges_0.5.4
[69] survival_3.5-5 iterators_1.0.14
[71] systemfonts_1.0.4 foreach_1.5.2
[73] tools_4.3.1 ragg_1.2.5
[75] sna_2.7-1 ica_1.0-3
[77] glue_1.6.2 gridExtra_2.3
[79] SparseArray_1.2.4 xfun_0.39
[81] MatrixGenerics_1.12.3 GenomeInfoDb_1.36.1
[83] withr_2.5.0 BiocManager_1.30.21.1
[85] fastmap_1.1.1 fansi_1.0.4
[87] callr_3.7.3 digest_0.6.33
[89] R6_2.5.1 mime_0.12
[91] textshaping_0.3.6 colorspace_2.1-0
[93] scattermore_1.2 tensor_1.5
[95] spatstat.data_3.0-1 RhpcBLASctl_0.23-42
[97] utf8_1.2.3 generics_0.1.3
[99] data.table_1.14.8 FNN_1.1.3.2
[101] httr_1.4.6 htmlwidgets_1.6.2
[103] S4Arrays_1.2.1 whisker_0.4.1
[105] uwot_0.1.16 pkgconfig_2.0.3
[107] gtable_0.3.3 registry_0.5-1
[109] ComplexHeatmap_2.16.0 lmtest_0.9-40
[111] SingleCellExperiment_1.22.0 XVector_0.40.0
[113] htmltools_0.5.5 carData_3.0-5
[115] dotCall64_1.1-1 clue_0.3-64
[117] scales_1.2.1 png_0.1-8
[119] knitr_1.43 rstudioapi_0.15.0
[121] rjson_0.2.21 reshape2_1.4.4
[123] coda_0.19-4 statnet.common_4.9.0
[125] nlme_3.1-162 cachem_1.0.8
[127] zoo_1.8-12 GlobalOptions_0.1.2
[129] stringr_1.5.0 KernSmooth_2.23-22
[131] parallel_4.3.1 miniUI_0.1.1.1
[133] pillar_1.9.0 grid_4.3.1
[135] vctrs_0.6.3 RANN_2.6.1
[137] ggpubr_0.6.0 promises_1.2.0.1
[139] car_3.1-2 xtable_1.8-4
[141] cluster_2.1.4 evaluate_0.21
[143] cli_3.6.1 compiler_4.3.1
[145] rlang_1.1.1 crayon_1.5.2
[147] rngtools_1.5.2 ggsignif_0.6.4
[149] future.apply_1.11.0 labeling_0.4.2
[151] ps_1.7.5 getPass_0.2-4
[153] plyr_1.8.8 fs_1.6.3
[155] stringi_1.7.12 network_1.18.1
[157] BiocParallel_1.34.2 viridisLite_0.4.2
[159] deldir_1.0-9 gridBase_0.4-7
[161] munsell_0.5.0 lazyeval_0.2.2
[163] spatstat.geom_3.2-4 Matrix_1.6-4
[165] patchwork_1.1.2 future_1.33.0
[167] shiny_1.7.4.1 highr_0.10
[169] SummarizedExperiment_1.30.2 ROCR_1.0-11
[171] broom_1.0.5 bslib_0.5.0 date()[1] "Wed Oct 1 17:31:41 2025"
sessionInfo()R version 4.3.1 (2023-06-16)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.6.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] bigmds_3.0.0 ggsci_3.0.0 harmony_1.2.0
[4] Rcpp_1.0.11 CellChat_1.6.1 Biobase_2.60.0
[7] BiocGenerics_0.46.0 igraph_1.5.0.1 dplyr_1.1.2
[10] dittoSeq_1.12.1 ggplot2_3.4.2 SeuratObject_5.1.0
[13] Seurat_4.3.0.1 purrr_1.0.1 here_1.0.1
[16] magrittr_2.0.3 circlize_0.4.15 tidyr_1.3.0
[19] tibble_3.2.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.21 splines_4.3.1
[3] later_1.3.1 bitops_1.0-7
[5] polyclip_1.10-4 ggnetwork_0.5.12
[7] lifecycle_1.0.3 rstatix_0.7.2
[9] doParallel_1.0.17 rprojroot_2.0.3
[11] globals_0.16.2 processx_3.8.2
[13] lattice_0.21-8 MASS_7.3-60
[15] backports_1.4.1 plotly_4.10.2
[17] sass_0.4.7 rmarkdown_2.23
[19] jquerylib_0.1.4 yaml_2.3.7
[21] httpuv_1.6.11 NMF_0.26
[23] sctransform_0.4.1 spam_2.10-0
[25] sp_2.0-0 spatstat.sparse_3.0-2
[27] reticulate_1.36.1 cowplot_1.1.1
[29] pbapply_1.7-2 RColorBrewer_1.1-3
[31] abind_1.4-5 zlibbioc_1.46.0
[33] Rtsne_0.16 GenomicRanges_1.52.0
[35] RCurl_1.98-1.12 pracma_2.4.4
[37] git2r_0.33.0 GenomeInfoDbData_1.2.10
[39] IRanges_2.34.1 S4Vectors_0.38.1
[41] svd_0.5.5 ggrepel_0.9.3
[43] irlba_2.3.5.1 listenv_0.9.0
[45] spatstat.utils_3.1-0 pheatmap_1.0.12
[47] RSpectra_0.16-1 goftest_1.2-3
[49] spatstat.random_3.1-5 fitdistrplus_1.1-11
[51] parallelly_1.36.0 svglite_2.1.1
[53] leiden_0.4.3 codetools_0.2-19
[55] DelayedArray_0.28.0 tidyselect_1.2.0
[57] shape_1.4.6 farver_2.1.1
[59] matrixStats_1.0.0 stats4_4.3.1
[61] spatstat.explore_3.2-1 jsonlite_1.8.7
[63] GetoptLong_1.0.5 BiocNeighbors_1.18.0
[65] ellipsis_0.3.2 progressr_0.13.0
[67] ggalluvial_0.12.5 ggridges_0.5.4
[69] survival_3.5-5 iterators_1.0.14
[71] systemfonts_1.0.4 foreach_1.5.2
[73] tools_4.3.1 ragg_1.2.5
[75] sna_2.7-1 ica_1.0-3
[77] glue_1.6.2 gridExtra_2.3
[79] SparseArray_1.2.4 xfun_0.39
[81] MatrixGenerics_1.12.3 GenomeInfoDb_1.36.1
[83] withr_2.5.0 BiocManager_1.30.21.1
[85] fastmap_1.1.1 fansi_1.0.4
[87] callr_3.7.3 digest_0.6.33
[89] R6_2.5.1 mime_0.12
[91] textshaping_0.3.6 colorspace_2.1-0
[93] scattermore_1.2 tensor_1.5
[95] spatstat.data_3.0-1 RhpcBLASctl_0.23-42
[97] utf8_1.2.3 generics_0.1.3
[99] data.table_1.14.8 FNN_1.1.3.2
[101] httr_1.4.6 htmlwidgets_1.6.2
[103] S4Arrays_1.2.1 whisker_0.4.1
[105] uwot_0.1.16 pkgconfig_2.0.3
[107] gtable_0.3.3 registry_0.5-1
[109] ComplexHeatmap_2.16.0 lmtest_0.9-40
[111] SingleCellExperiment_1.22.0 XVector_0.40.0
[113] htmltools_0.5.5 carData_3.0-5
[115] dotCall64_1.1-1 clue_0.3-64
[117] scales_1.2.1 png_0.1-8
[119] knitr_1.43 rstudioapi_0.15.0
[121] rjson_0.2.21 reshape2_1.4.4
[123] coda_0.19-4 statnet.common_4.9.0
[125] nlme_3.1-162 cachem_1.0.8
[127] zoo_1.8-12 GlobalOptions_0.1.2
[129] stringr_1.5.0 KernSmooth_2.23-22
[131] parallel_4.3.1 miniUI_0.1.1.1
[133] pillar_1.9.0 grid_4.3.1
[135] vctrs_0.6.3 RANN_2.6.1
[137] ggpubr_0.6.0 promises_1.2.0.1
[139] car_3.1-2 xtable_1.8-4
[141] cluster_2.1.4 evaluate_0.21
[143] cli_3.6.1 compiler_4.3.1
[145] rlang_1.1.1 crayon_1.5.2
[147] rngtools_1.5.2 ggsignif_0.6.4
[149] future.apply_1.11.0 labeling_0.4.2
[151] ps_1.7.5 getPass_0.2-4
[153] plyr_1.8.8 fs_1.6.3
[155] stringi_1.7.12 network_1.18.1
[157] BiocParallel_1.34.2 viridisLite_0.4.2
[159] deldir_1.0-9 gridBase_0.4-7
[161] munsell_0.5.0 lazyeval_0.2.2
[163] spatstat.geom_3.2-4 Matrix_1.6-4
[165] patchwork_1.1.2 future_1.33.0
[167] shiny_1.7.4.1 highr_0.10
[169] SummarizedExperiment_1.30.2 ROCR_1.0-11
[171] broom_1.0.5 bslib_0.5.0